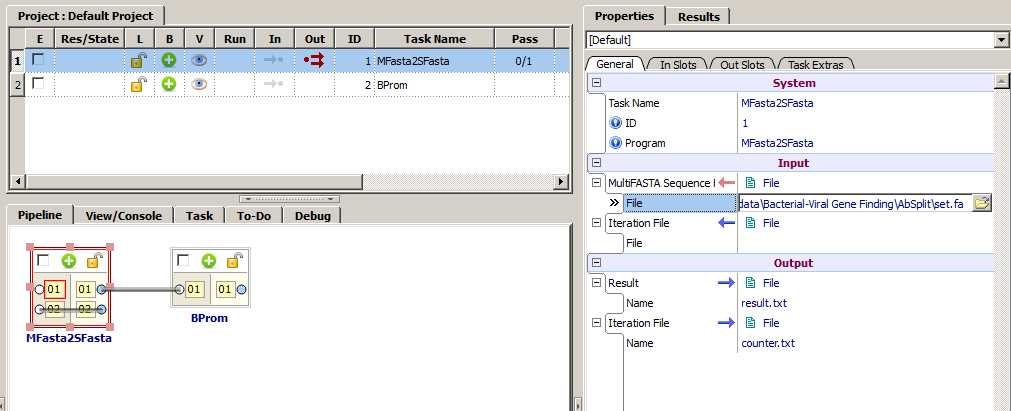
MFasta2SFasta serves to split multifasta files into singlefasta ones.
Current version of MolQuest does not support multifasta files for BPROM input. However, it is possible to process multifasta files via the following sequence of procedures: split multifasta files into singlefasta ones with use of the MFasta2SFasta utility and then send them to BPROM via pipelines. This requires the following steps.
Add to project MFasta2SFasta and BPROM. It requires to open the "Toolbox>Programs" item and double click on appropriate program/utility. MFasta2SFasta can be found in the "All programs" and "SeqMan" list sections, BPROM – in the "All programs" and "Eukaryotic gene Finding" ones.
In setup of of MFasta2SFasta do the following:

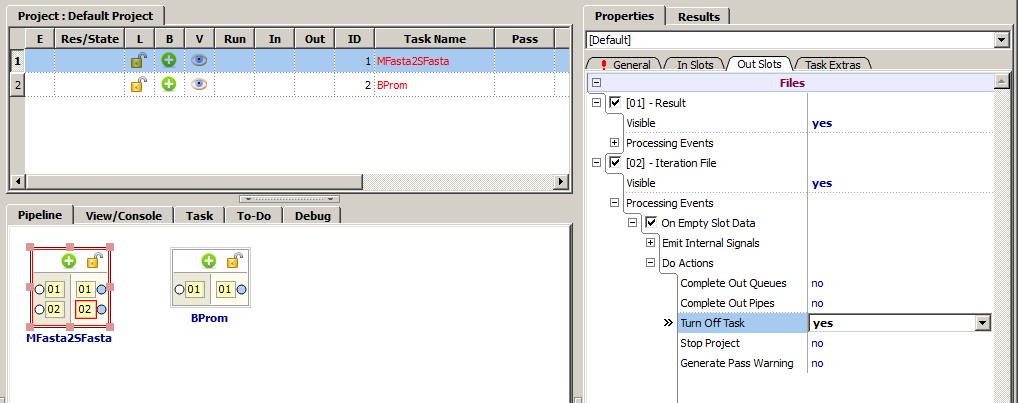
1. Find the "Iteration File" parameter (can be found in the "Properties" tab, in the "Out Slots" subtab), and in the "Processing Events" node check in the "On Empty Slot Data" check-box and further in the "Do Actions" subnode set the "Turn Off Task" value to "yes".
2. Link the output "[02] - Iteration File" to its input "[02] - Iteration File" (can be found in the "Properties" tab, in the "In Slots" subtab), i.e. to cycle it to itself.
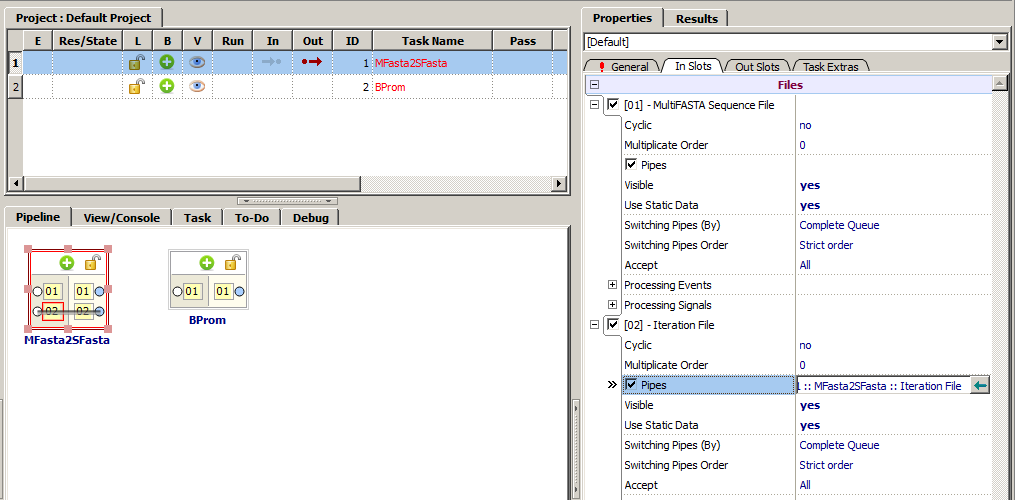
For MFasta2SFasta do the following:
1. To the slot "MultiFASTA Sequence File" (can be found in the "Properties" tab, in the "General" subtab) send a file with sequences (multifasta).
2. The first out slot "1 :: MFasta2SFasta :: Result" of the MFasta2SFasta link with the in slot "Sequences set" ("01") of the BPROM.